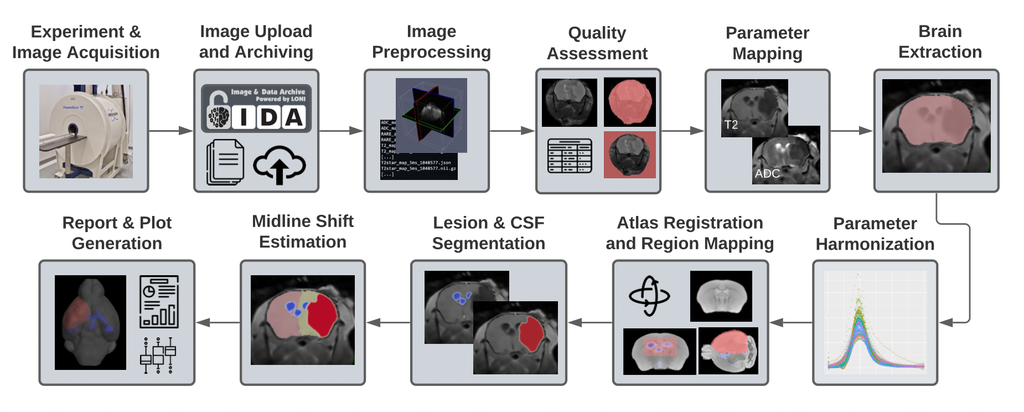
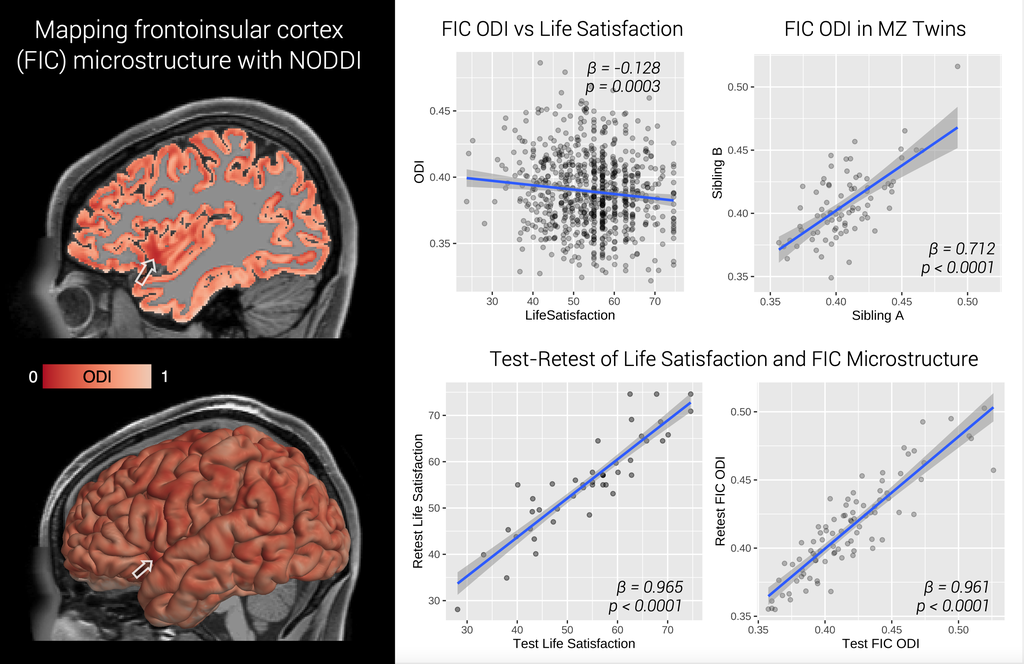
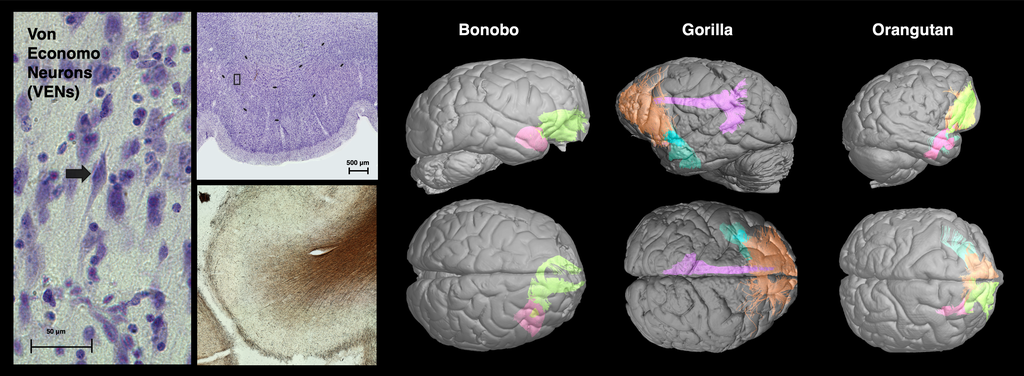
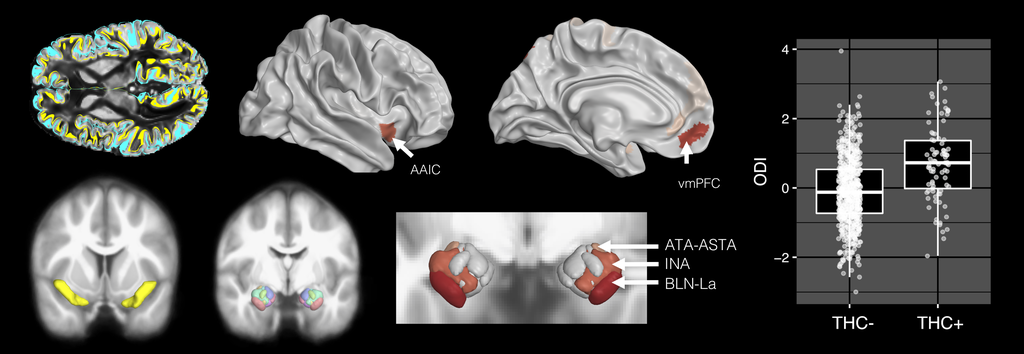
Welcome! I am a computational imaging researcher and software developer dedicated to improving the capabilities and impact of biomedical imaging technology. I am currently a Staff Machine Learning Engineer at ArteraAI, where I am working on computer vision for AI-enabled predictive and prognostic cancer tests. Additionally, I am an Adjunct Assistant Professor of Research at the Stevens Neuroimaging and Informatics Institute (INI) at the University of Southern California (USC).
Prior to this, I conducted academic research in computer science and neuroscience at the USC Laboratory of Neuro Imaging with generous support from the Chan Zuckerberg Imaging Scientist program. I hold a PhD in Computer Science from Brown University, where I was advised by David H. Laidlaw, and a BSc from the California Institute of Technology. My research has centered on developing computational methods and software for characterizing the brains of humans and rodents using mathematical modeling, 3D visualization, and machine learning. Through interdisciplinary collaborations, we have applied these techniques to map normative changes throughout development and aging, as well as disruptions associated with dementia, environmental exposure, traumatic brain injury, and stroke.
I invite you to learn more about my work by exploring my publications, some of which are listed below. Much of my research utilizes freely-available software that I developed, known as the Quantitative Imaging Toolkit (QIT). You can learn more about QIT by visiting its dedicated website.
Interests
- Machine Learning
- Biomedical imaging
- Scientific Visualization
- Computational Image Analysis
- Software Development
Education
-
PhD in Computer Science, 2016
Brown University
-
MSc in Computer Science, 2012
Brown University
-
BSc in Eng. & Applied Science, 2005
California Institute of Technology